Systems Biology of Tumor Immune Stromal Interactions in Metastatic Progression
Although immunotherapy has improved treatment of metastatic cancer, not all patients respond, and new treatment strategies are needed. We hypothesize a novel mechanism for metastasis: that it is prompted by interactions between tumor cells, immune cells, and stromal cells inside lymph nodes. This concept is a paradigm shift that could alter our fundamental understanding of cancer spread as well as point to new treatments.
Our CCSB Research Center interweaves mouse models and primary human data in head and neck cancer and lung cancer. We analyze highly multiplexed, multi-scale datasets with novel bio-computational methods to reconstruct intracellular and intercellular crosstalk between cell types in the microenvironment of primary tumors, lymph nodes, and distant metastases. This research is organized around two projects and one shared core that are closely integrated.
Mouse models analysis of lymph node and distant metastasis
We hypothesize that local interactions between tumor cells, leukocytes, and stroma within lymph nodes initiate the metastatic cascade through lymph node tolerization. Using syngeneic mouse models, we will identify the mechanisms for lymph node tolerization.
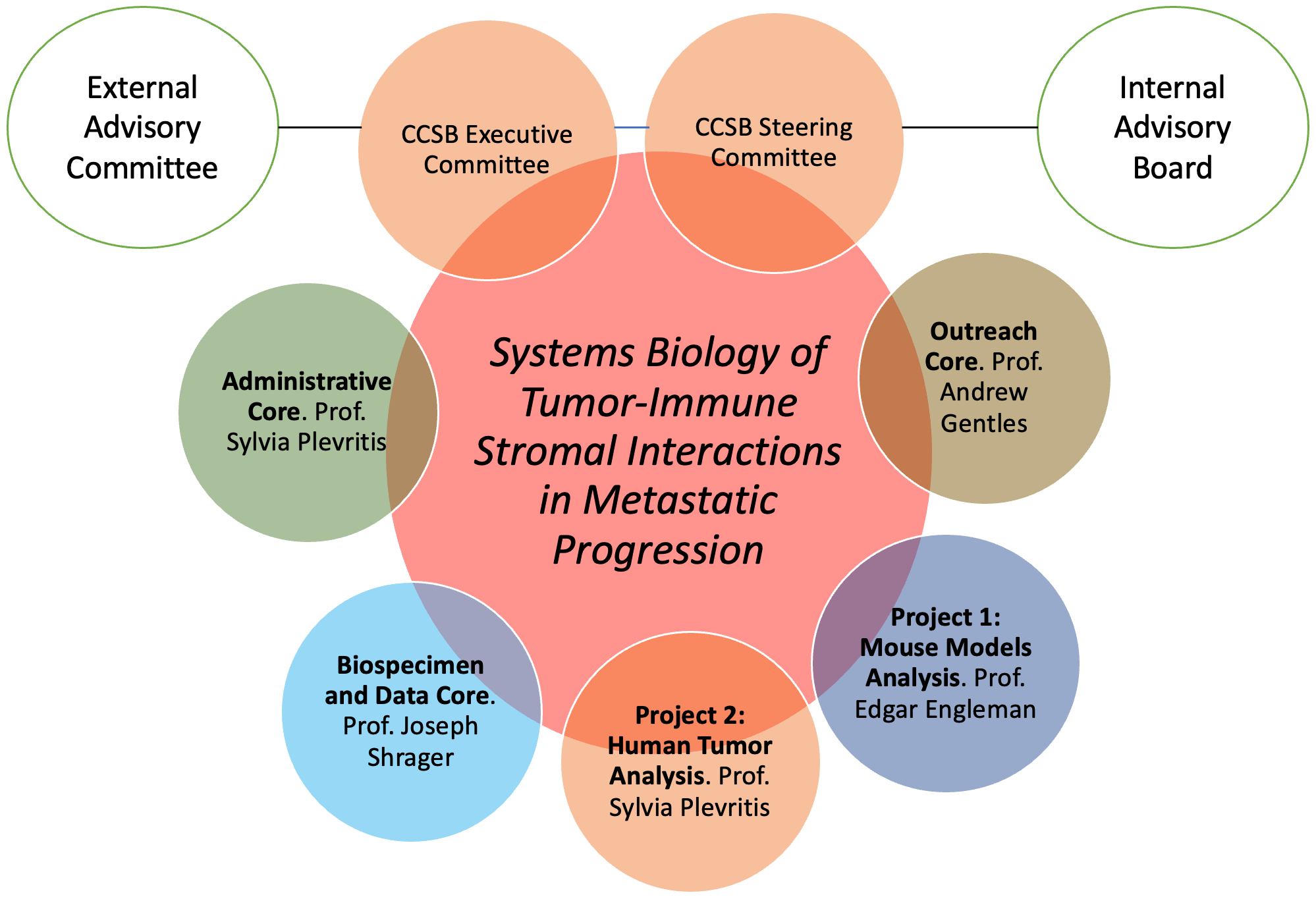
Organizational Structure of the Stanford Center of Cancer Systems Biology
To learn more, please visit the Cancer Systems Biology Consortium website